Help
This is the user manual for the website.
1 GreenCells Homepage
The homepage of the GreenCells database displays a general navigation bar, Introduction, Quick Search, and Plant Browse. GreenCells provides 6 main functional modules, including Browse, Search, Tools, Statistics, Download, and Help.

2 Browse
The Browse drop-down contains a selection of 8 species. Users can click on a species of interest to access a comprehensive information to browse, including Overview, Introduction, Genomic Information, Tissue, Project. Furthermore, by clicking the view icon located on the right side, users can directly navigate to the detailed information page of a specific Project or Tissue they find interesting.
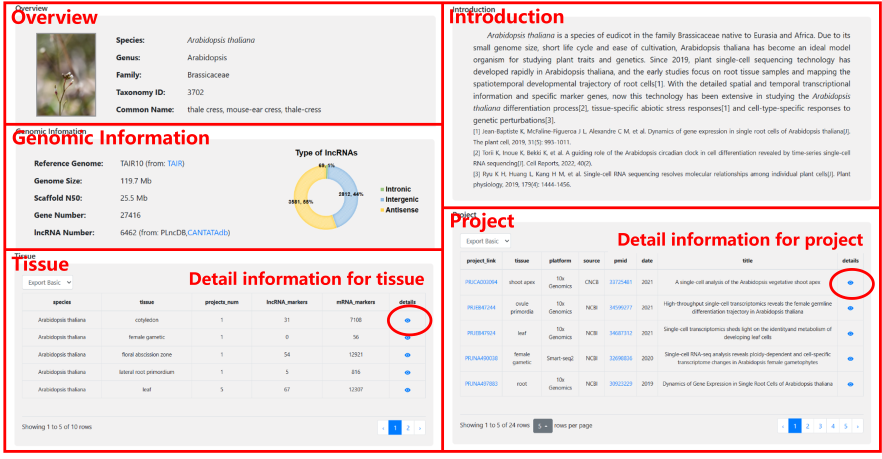
On the detailed information page of each project, users can view the following:
Basic Info: This section provides an overview of the project, including its source, species, tissues, platform, summary, submissions and other detail information.

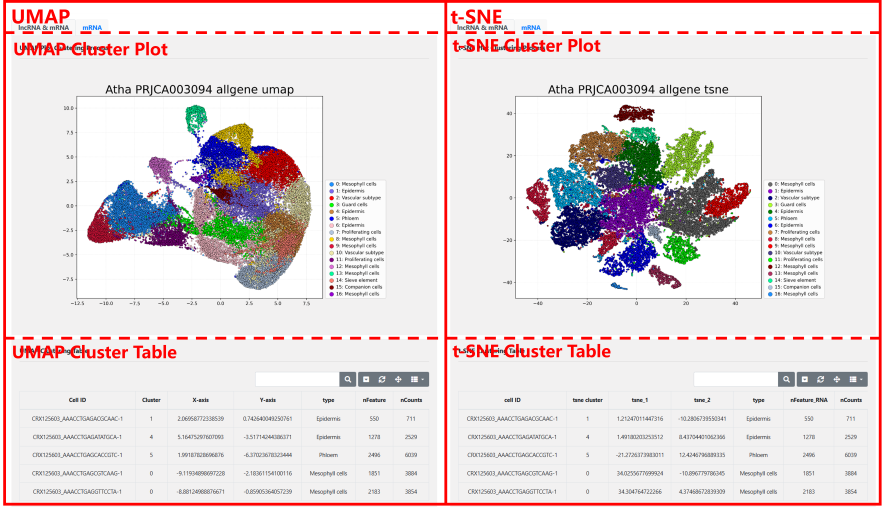
UMAP Visualization: The UMAP diagram shows the clustering and annotation information using the UMAP algorithm.
t-SNE Visualization: The t-SNE diagram shows the clustering and annotation information using the t-SNE algorithm.
Marker Genes: This section displays the top 10 marker genes in each cell cluster predicted by Seurat’s FindAllMarkes function. You can review the list of marker genes to learn more about these genes and their characteristics.

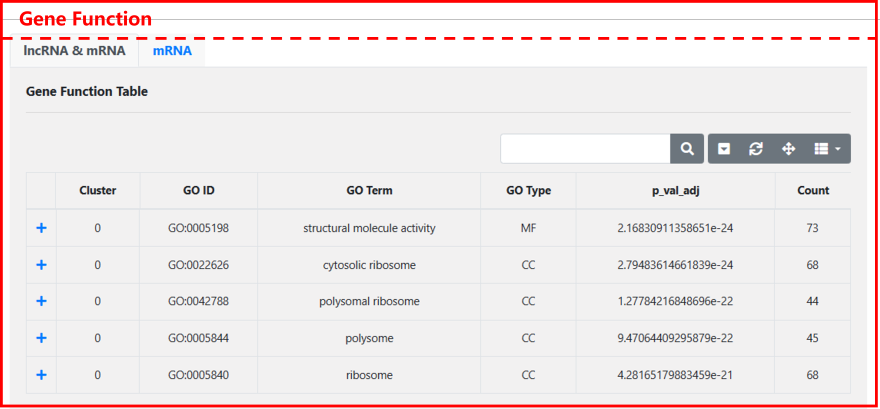
Gene Function: We conduct gene function enrichment analysis for each cell cluster, providing enriched genes along with their corresponding GO terms. This allows for a deeper understanding of the function of specific cell clusters and facilitates further exploration of these information.
Cell-cell Communication: We inferred the cell-cell communication across each cell-type, and we display the chord diagram of the results, and the receptor and ligand respondence genes are displayed as table form.
Cell-type Network: This section provides the cell-type co-expression network outcomes, and display the top KME gene in each module within one cell-type as well as the dot plot that illuminate the expression pattern of module genes.

By clicking on different tabs or navigation options, you can switch between and explore different sections within the detailed information page. You should notice that the “project” section means the data come from only one project, and the “tissue” section integrate the same tissue samples datasets if it have more than one dataset. The results generated by the two buttons under the sub navigation stem from different analytical methods. "mRNA" denotes variable genes comprising only mRNA for downstream analysis. "lncRNA & mRNA" denotes variable genes comprising both lncRNA and mRNA for downstream analysis. Clicking on each button allows browsing through the respective results.
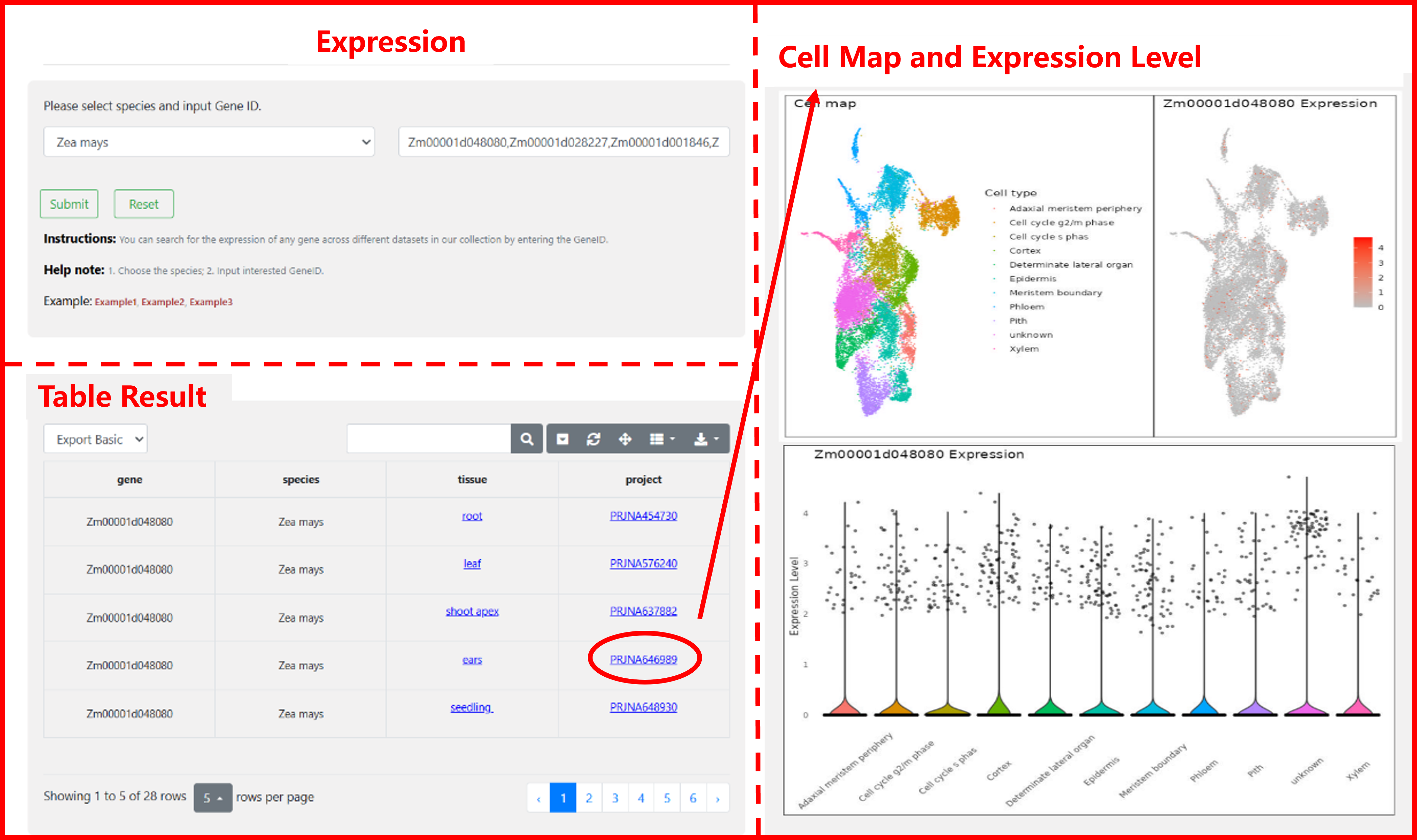
3 Expression
In this module, we provide the expression of any lncRNA/gene across different datasets in our collection. Users can choose the species, enter interested Gene, and you will obtain a result form. By clicking on the corresponding "Tissue" or "Project", you can view the cell map and expression level.
4 Search
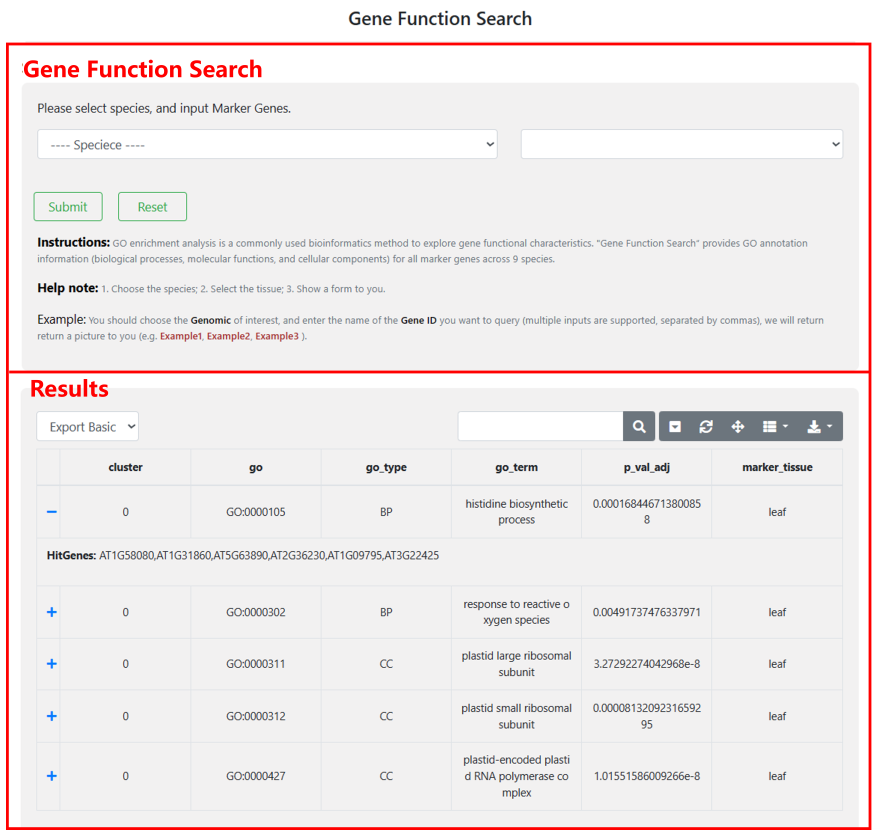
4.1 Gene Function Search
“Gene Function Search" provides the GO annotation of marker protein genes for each cell cluster in 8 species, covering a total of 14 tissue types. Users can choose the interested species, select the tissue, and you will obtain a result form including the information of GO enrichment.
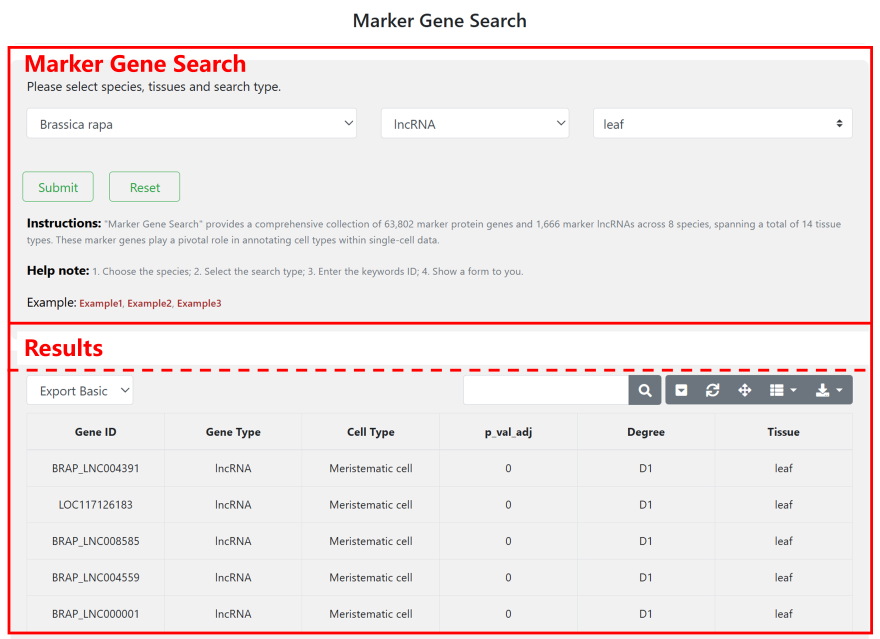
4.2 Marker Gene Search
"Marker Gene Search" offers an extensive collection of 63,802 marker protein genes and 1,666 marker lncRNAs across 8 species, covering 14 different tissue types. Users can select the species and gene type to retrieve a comprehensive gene table. The search box above the table allows users to find specific genes, tissues, or other terms. Please note that genes marked as "D1" represent high-confidence marker genes. These genes have an expression ratio greater than 0.25 within a specific cell group and an expression ratio within that cell group that is more than 2.5 times higher than their expression ratio in all cells. You can quickly download these files from the "Download" module of this website.
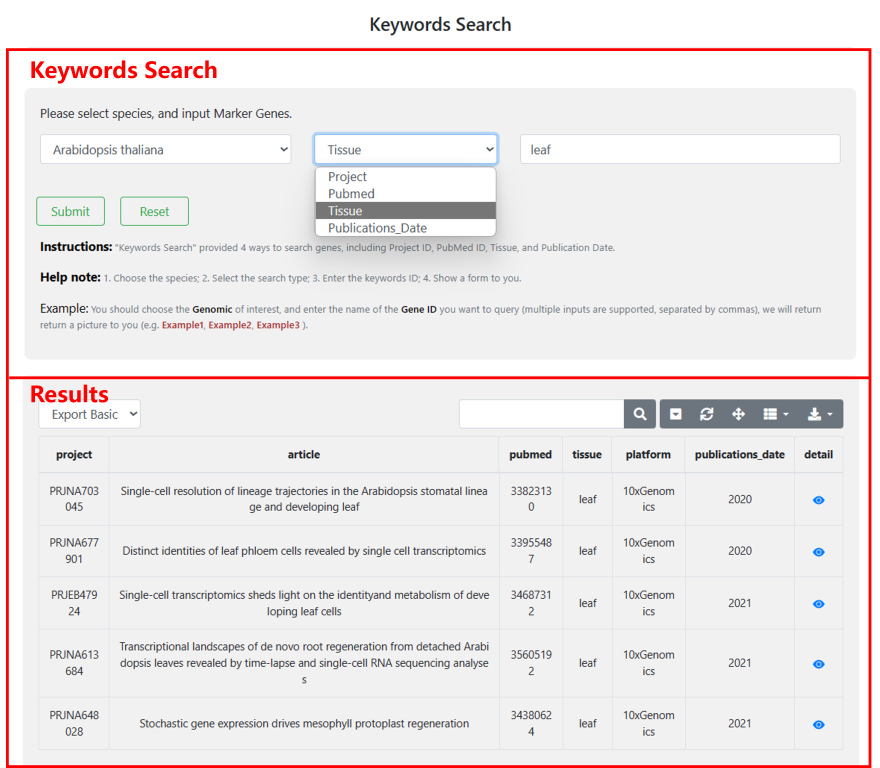
4.3 Keywords Search
"Keywords Search" provided 4 ways to search this database, including Project ID, PubMed ID, Tissue, and Publication Date. Users can choose the interested species, select the search type, enter the keywords ID, and then you will obtain a result form including the information of Project.
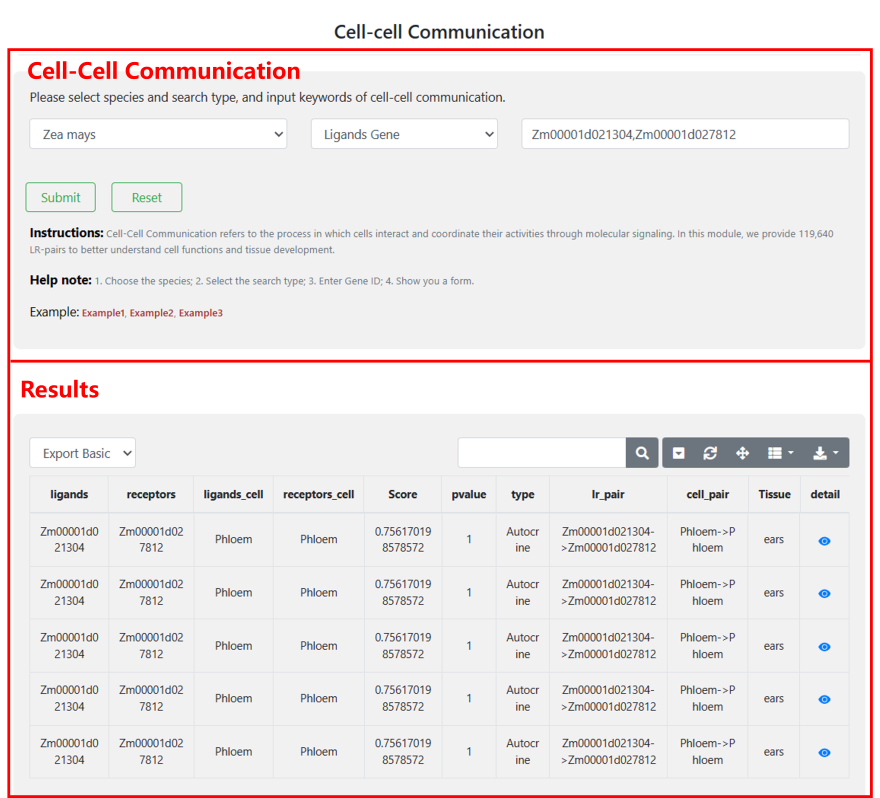
4.4 Cell-Cell Communication
“Cell-Cell Communication" offers 119,640 LR-pairs to enhance the understanding of interactions between different cell types. Users can select their species of interest, choose the search type, and enter a Gene ID to obtain a detailed result form. Please note that cell-cell communication data is available for only four species due to the limited availability of receptor-ligand databases.
5 Tools
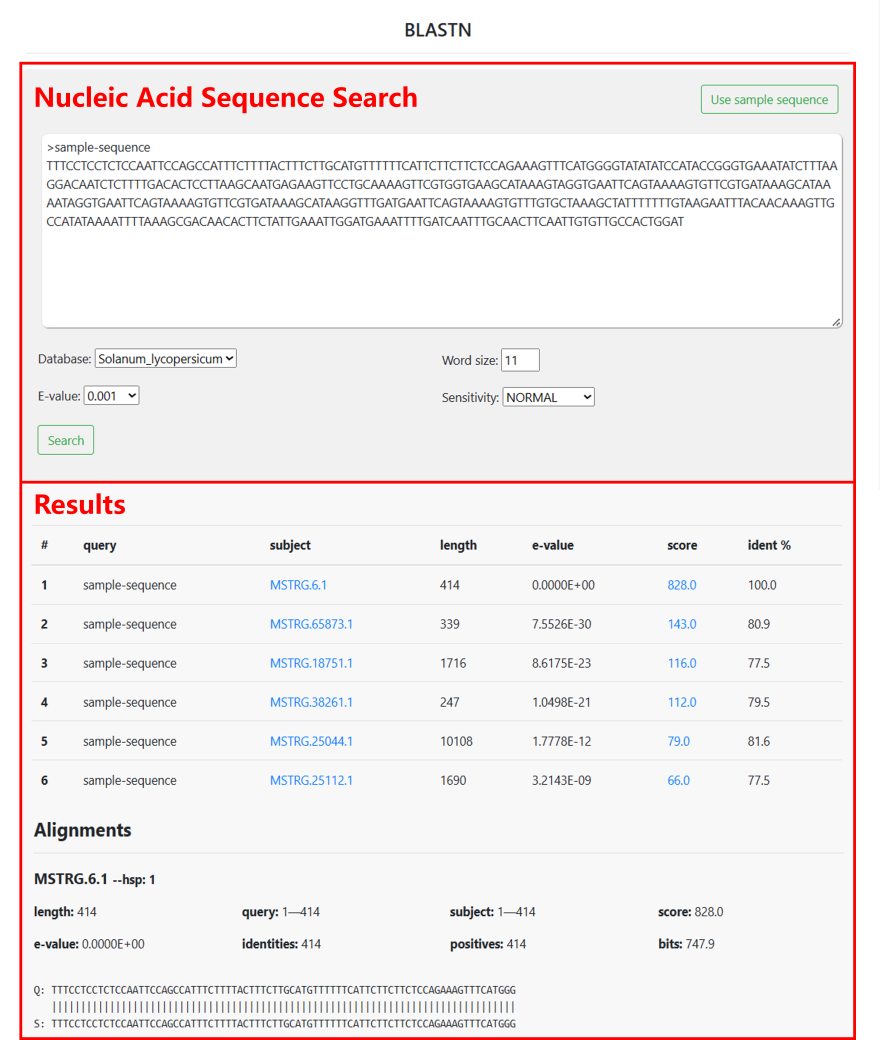
5.1 BLAST
GreenCells provides BLAST tools for sequence alignment of genes from 8 species within the site. Users can enter nucleic acid sequences, select the species and gene type of interest, set parameters, and then obtain alignment results.
5.2 CSN
The tool takes the original single-cell gene expression matrix as input and generates a network degree matrix (NDM). NDM, with dimensions mirroring those of the original gene expression matrix (GEM), prioritizes gene significance within the network over expression levels, ensuring compatibility with traditional scRNA-seq algorithms. Users can upload files (<5M) and set operating parameters to perform CSN online analysis, and then you will obtain a result file. If your file is large or you want to run it in the terminal, we provide a python script, you can run the command: python cns.py yourfile.

4.3 Plant scRNA Upstream analysis
In order to facilitate single-cell data analysis, we provide pipeline for 10x Genomic and Smart-seq.
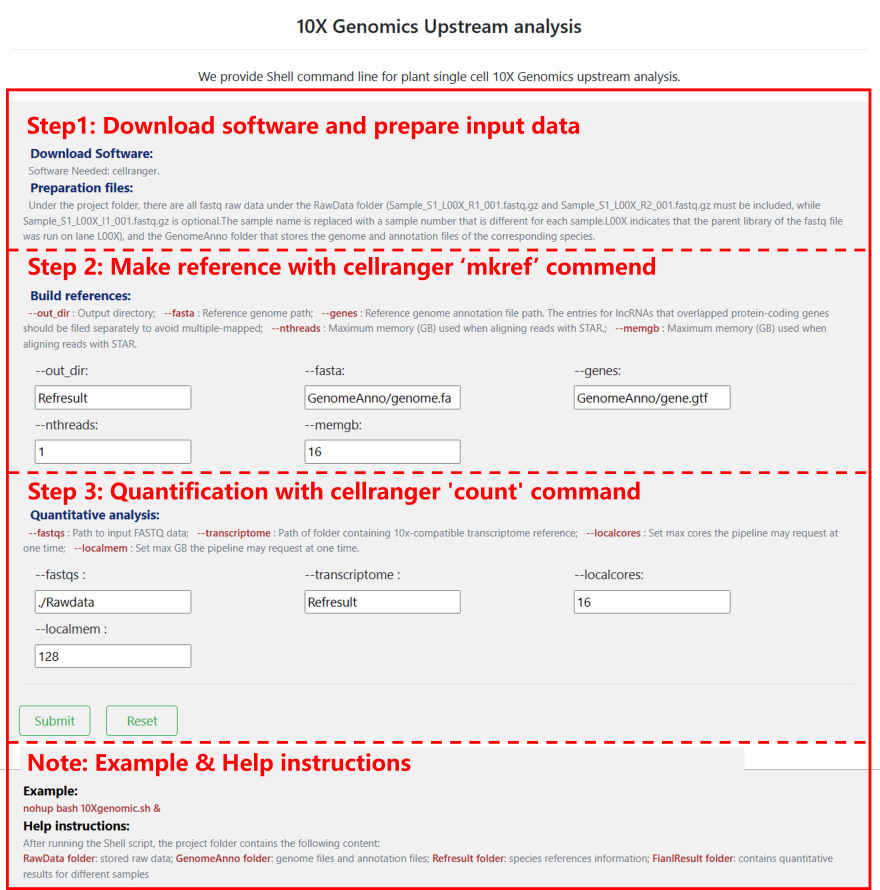
10x Genomics: This tool facilitates the processing of raw data generated by the 10x Genomics protocol, enabling the transformation of fastq files into an expression count matrix. Users can set parameters as needed and obtain a runnable shell script. Additionally, the necessary tools and files must be prepared according to the tool's instructions.
Smart-seq: This tool facilitates the processing of raw data generated by the 10x Genomics protocol, enabling the transformation of fastq files into an expression count matrix. Users can set parameters as needed and obtain a runnable shell script. Additionally, the necessary tools and files must be prepared according to the tool's instructions.

4.4 Plant scRNA Downstream analysis
We provide downstream analysis tools suitable for data from 10x Genomics and Smart-seq. First, install the required software and packages as instructed, and set the personalized parameters. You will then receive a downstream analysis script that can be executed in the terminal. Please note that the marker genes used for cell annotation must adhere to the specified file format. Marker genes for various plant species can be downloaded from our website.

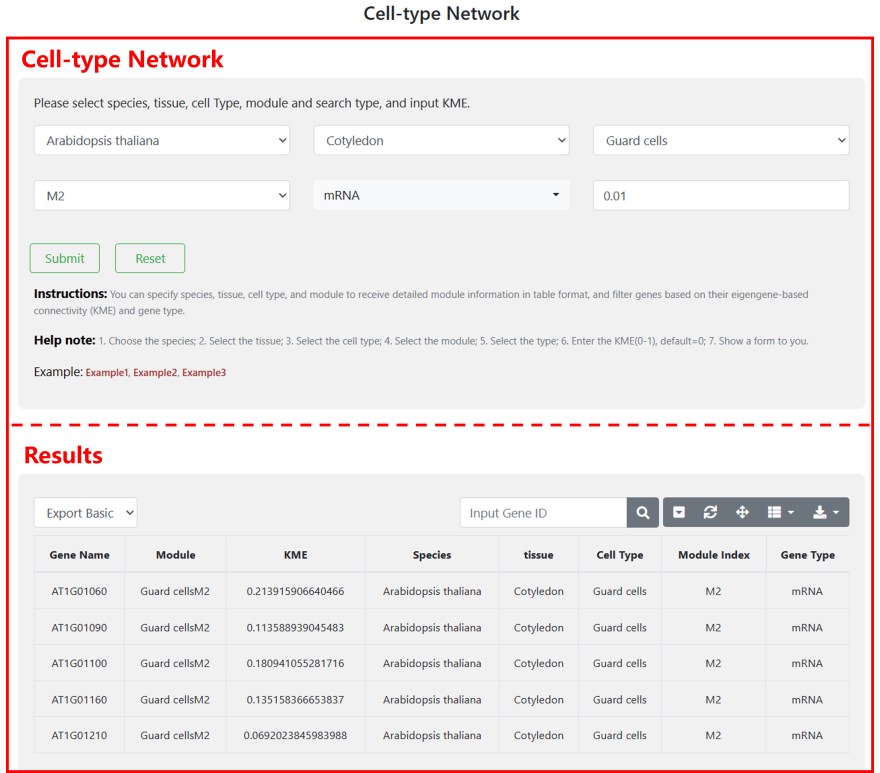
4.5 Cell-Type Network
Users can specify their desired species, tissues, cell types, and modules of interest. Then tool returns module information within the network, including genes contained within each module, the eigengene-based connectivity coefficient for each gene, and annotations indicating gene types. Additionally, users have the option to filter out genes with lower connectivity coefficients within the modules based on their preferences, as exploring the functionality of genes holds paramount significance, we propose further investigation of lncRNAs through coding genes within modules.
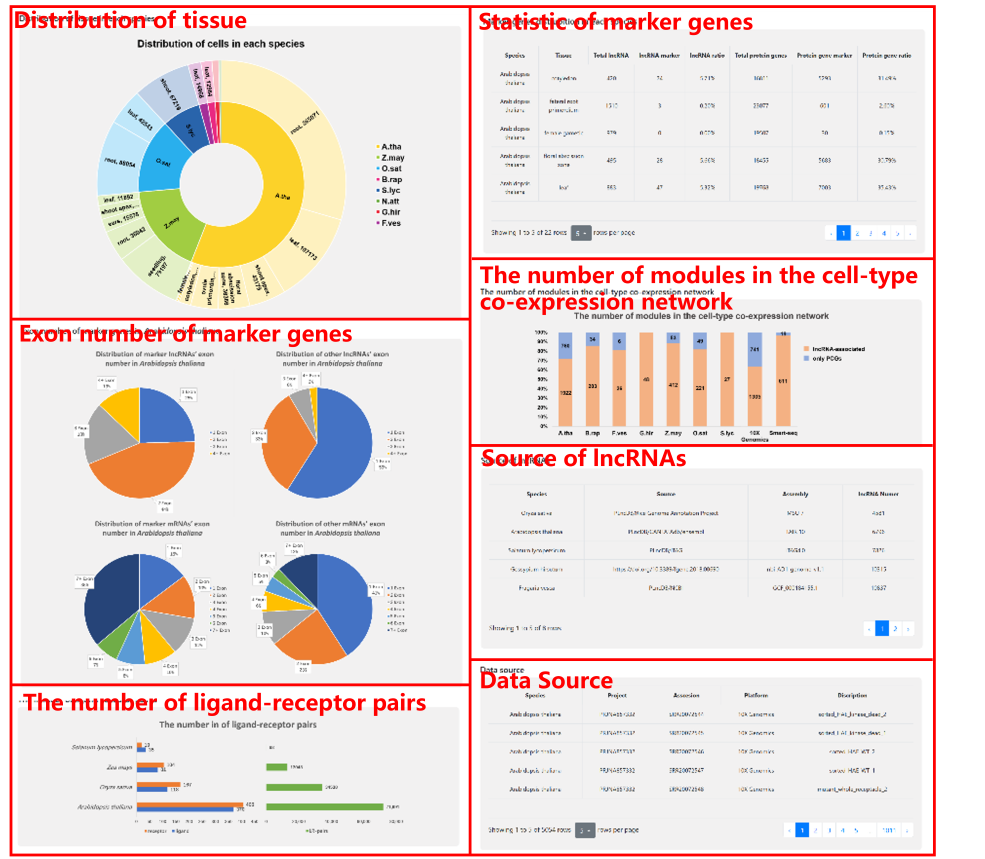
6 Statistics
We provide statistics of Distribution of cell in each species, Marker genes distrubition in each species, The number of ligand-receptor paires, The number of modules in the cell-type co-expression network, Exon number of marker genes in Arabidopsis thaliana, Source of lncRNAs, and Data source.
7 Download
We provide Annotation Download, Novel Marker Genes Download, and Known Marker Genes Download.